Tutorial for Circ2Disease
1. Overview of Circ2Disease
Circ2Disease is a database that intergrated a comprehensive collection of
human circRNA and disease associations. All those associations were manually
retriewed from public literature. We also predicted the corresponding miRNA
sponges of those validated circRNAs and integrated RBP informaiton from
CircInteratome database. Then experimentally verified miRNA-disease
associations and miRNA targets were united from several public databases.
All those data formed a large regulation network to provide more information
for users.
Now the database can provide browsing, searching, network analysis and downloading
functions. See below for more details.
2. Database Construction
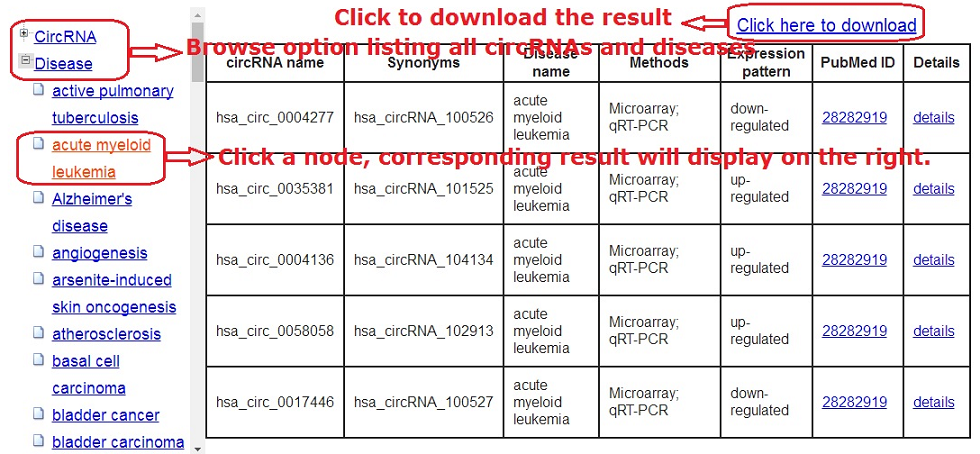
3. Browse function
The Browse page for general perusal of the database based on different
hierarchical classification of circRNAs and diseases trees.
Selecting particular nodes will display a corresponding result table.
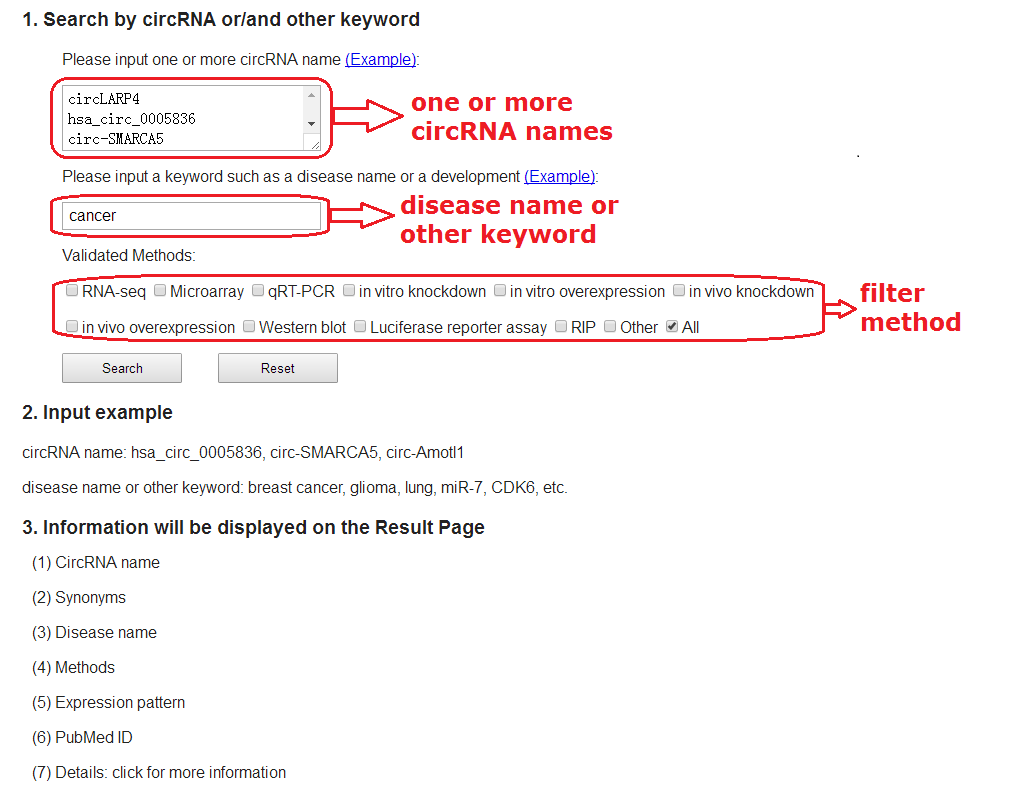
4. Search function
The Search page allows users to input one or more circRNA name, disease name or other
keyword to query related associations in Circ2Disease. In addition, users can specify
the verified experimental methods to filter the result.
5. CircRNA general information
When users initiate a search request, a tips panel displaying the search details
appears at the top of the results page. Circ2Disease results are organized in a
data table, with a single association record on each line that contains circRNA
name, diseas name, expression pattern and PubMed ID. Users can click on
the "click here to download" to download the data table and the "details"
link to view detailed information of the particular association. All the detailed
information are included in the download data table not only the gereral information.

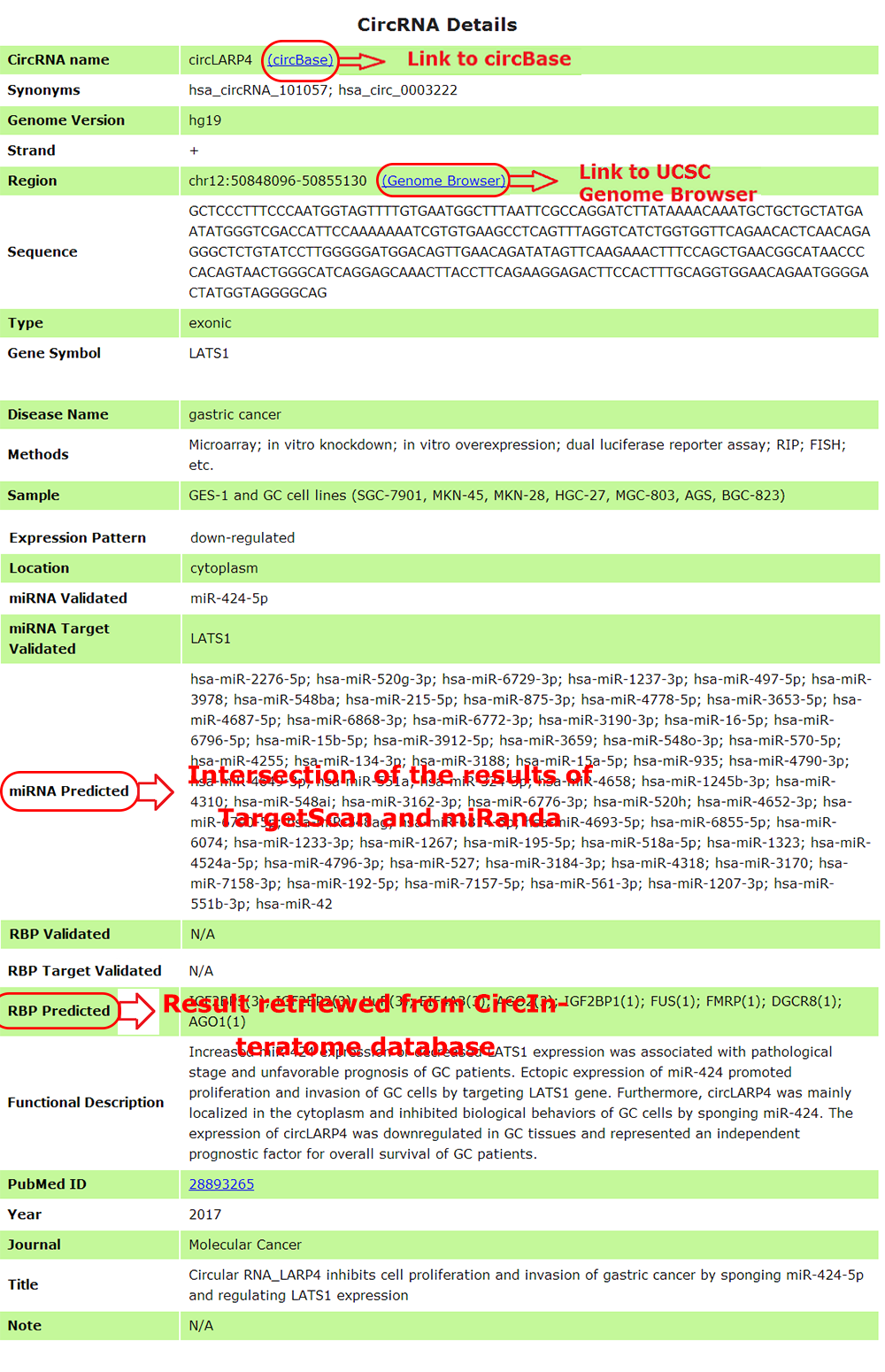
6. CircRNA details
Users can obtain detailed information in Circ2Disease by clicking on the "details"
of the data table, which will open a page containing comprehensive information
of the single record and provides links to external web sites, such as
circBase and Genome Browser.Provides genomic annotation of circRNAs with a genome
browser integrated into the web interface.
7. Network analysis
(1) CircRNA network
Validated circRNA-disease interactions were retriwed from literature.If any
circRNA-miRNA sponge, circRNA-RBP interactions, miRNA target, and upstream or
downstream genes were also extracted to generate circRNA regulatory network.
This network page can be found here:
circRNA_network page.
This network will display as below:
The network page supports the following functions:
- Search: circRNA, miRNA, disease and gene can be input to search the network and is case-insensitive. The matched node will turn yellow and display in the center of the page.
- Zoom: Scroll the mouse wheel to zoom in and out of the network.
- Display: Grab and drag nodes; tap a node or edge to select; tap blank to unselect.
- Hide/Show: The selected nodes and edges will be hidden or shown.
- Neighbor: Click the "Neighbor" button to select nodes and edges connected to the previously selected node.
- Export: Export the displayed network to JPG or PNG file. You can save it on your local computer.
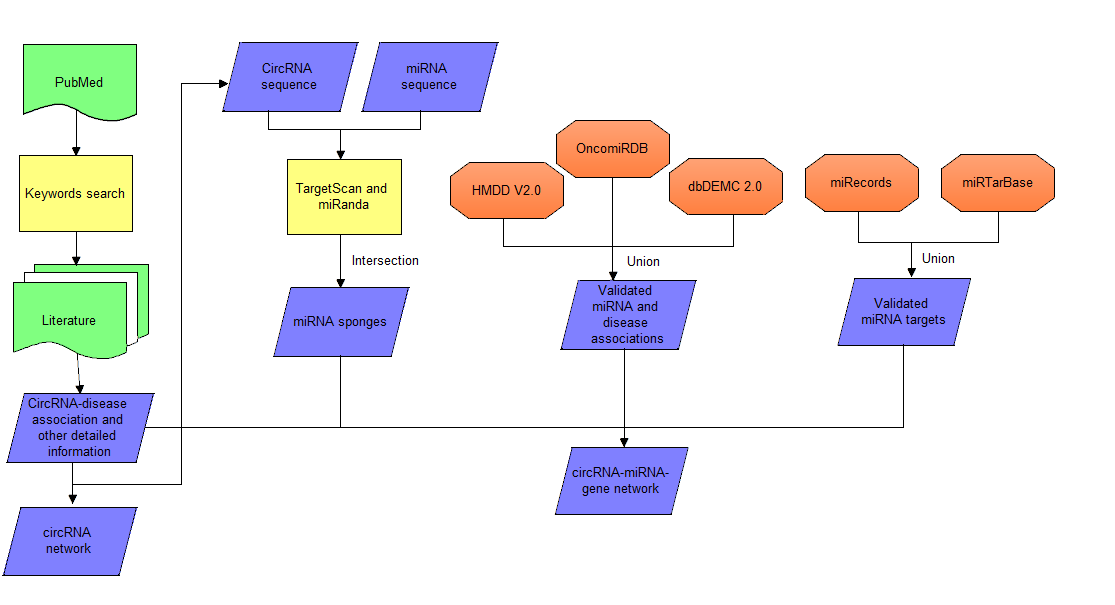
(2) CircRNA-miRNA-gene network
MicroRNA Sponge Detection: Potential miRNA binding sites on circRNAs were identified by targetscan
and miRanda. We seperately ran TargetScan with default parameters and miRanda with the "strict" parameter.
To increase the specificity, 6mer matches were excluded from the TargetScan result.
The intersection of the two results were considered as the predicted miRNA sponge.
To investigate the association between those miRNAs, their targets and diseases,
we integrated several public databases, i.e. three experimentally validated miRNA
databases (HMDD v2.0, dbDEMC 2.0 and OncomiRDB) and two experimentally validated
miRNA-target databases (miRTarBase and miRecords). For data from miRTarBase, only
strong experimental evidence of MTIs (miRNA–target interactions) were retained
for further network analysis.
All these interactions formed a comprehesive circRNA-miRNA-gene regulatory
network, which will help users to
elucidate the pathogenesis of diseases and offer beneficial evidence for
diagnosis and therapy.
8. Download
Users can download all data in Circ2Disease from "Download" page, including:
- The experimentally validated circRNA-disease association data.
- The manually curated circRNA regulatory network derived from literature.
- CircRNA name and alias.
- CircRNA related diseases from literature.
- All diseases association with circRNAs and miRNAs.
- Predicted RBP interacting with circRNA from CircInteractome database.
- The experimentally verified miRNA-disease association data integrated from HMDD, OncomiRDB and dbDEMC databases.
- The experimentally verified miRNA target data integrated from miRecords and miRTarBase databases.
9. Help information
The "Help" drop-down menu displays tutorial and information about how to
use Circ2Disease, containing circRNA alias and disease names in orignal paper
and normalized name.
10. Contact us
Liu Pengyuan's Lab:
http://bioinformatics.zju.edu.cn. For any questions or
suggestions about our website, please
contact us: dxyao1984@163.com,
and pyliu@zju.edu.cn.
